In molecular biology, an interactome is the whole set of molecular interactions in a particular cell. Significance Analysis of INTeractome (SAINT) consists of a series of software tools for assigning confidence scores to protein-protein interactions based on quantitative proteomics data in AP-MS experiments. Submit your interactions and the server will find all the available structural data for both the single interactors and the interactions themselves. The interactome is the totality of PPIs that happen in a cell, an organism or a specific biological context. The development of large-scale PPI screening techniques, especially high-throughput affinity purification combined with mass-spectrometry and the yeast two-hybrid assay, has caused an explosion in the amount of PPI data.
Are all interactome types interconnected? What is the topology of an interactome? CCSB interactome mapping and ORFeome cloning efforts are supported by federal grants from the National Human Genome Research Institute, the National Cancer Institute and the National Science Foundation, and by funding from the Dana-Farber Cancer Institute Strategic Initiative, The Ellison Foundation and the W. H uman interactome mapping is the flagship project of CCSB. Interactome Projects at CCSB. Its clever node technology engages visitors in a way that has them interacting with your videos.
The B cell interactome (BCI) is a network of protein-protein, protein-DNA and modulatory interactions in human B cells. The network contains known interactions (reported in public databases) and predicted interactions by a Bayesian evidence integration framework which integrates a variety of generic and context specific experimental clues about protein-protein and protein. The advantage of the precision-recall curve is that it shows precision and recall rates on different cut-offs, which gives a more comprehensive view of the quality of the interactome , independent of the selection of cut-offs. The higher the AUC, the better an interactome may support ‘guilt-by-association’ prediction of gene function.

Underlying this tool is the application of machine learning to predict protein interaction interfaces for 189protein interactions with previously unresolved interfaces in human and seven model organisms, including the. Sangerman et al reported the association of ubiquitin with SPTAN(αSpII) in neurons, 9but this association does not appear in the interactome database. Cytoscape is an open source software platform for visualizing molecular interaction networks and biological pathways and integrating these networks with annotations, gene expression profiles and other state data. SmartTMT Software Real-Time Search. This is an important point.
Time consuming SPS-MS spectra are only acquired after confident peptide identification, greatly increasing the number of peptides interrogated and reducing the. Our research encompasses the areas of bioinformatics and software development, methods development and biological applications. The integration of all the elements in the proteomics pipeline within one lab facilitates advances in all of them. The Yates lab has published more than 7peer reviewed papers. Validation in a benchmarked in vitro pull-down assay revealed that a random subset of TF-NAPPA validated at the same rate of as a positive reference set of literature-curated interactions.
The SIB Swiss Institute of Bioinformatics is happy to announce the new version of Expasy on October, with a brand new user interface, reviewed resource annotations, and a new visual identity! In the decade since then, genetic studies have revealed at least dominant mutations in LRRKlinked to Parkinson’s disease, alongside one associated with cancer. Here, we report on an interactome map that focuses on neurodegenerative disease (ND), connects ∼0human proteins via ∼30candidate interactions and is generated by systematic yeast two-hybrid interaction screening of ∼5ND-related proteins and integration of literature interactions. Mitochondrial fractionation.
As a result, the later years of life are often spent in poor health and lowered quality of life. Revealing the human-microbial interactome will allow further understanding of the mechanisms behind the onset of oral diseases. Additionally, this knowledge may give insight on key proteins involved in oral infections, which can be used for either diagnosis, as molecular biomarkers, or for treatment, as drug-targets. KinomeNetworkX A Network Atlas of 5Human Kinase genes. DESnowball Method to identify downstream target genes that are affected by a recurrent driver mutation.
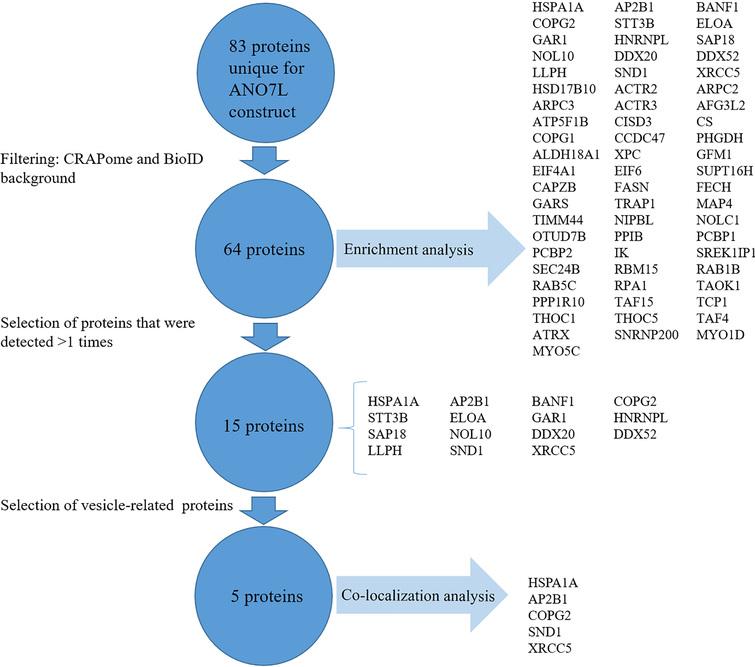
PAHKB Pulmonary Arterial Hypertension KnowledgeBase. Semi-supervised SAINT models with controls are used for all experiments in this interaction repository. However, this was a software -imposed threshold. In summary, the interactome analysis aided to guide the design of novel models of SARS-CoV pathogenicity. Although the legume–rhizobium symbiosis is a most-important biological process, there is a limited knowledge about the protein interaction network between host and symbiont.
Pathway Commons - Pathway Commons is a collection of publicly available pathways from multiple organisms. It provides researchers with convenient access to a comprehensive collection of pathways from multiple sources represented in a common language. Nucleic Acids Research. For network analysis, high-confidence CAR interactions (SaintScore, ) and substantially altered phosphosites were used to generate a network (STRING). Your article has been reviewed by three peer reviewers, and the evaluation has been overseen by a Reviewing Editor and a Senior Editor.

Welcome to interacto. To use this tool, first, choose your organism of interest. Then, push the corresponding button at the top left, to generate the protein-protein interaction networks for that organism. Reactome is a free, open-source, curated and peer-reviewed pathway database.
Our goal is to provide intuitive bioinformatics tools for the visualization, interpretation and analysis of pathway knowledge to support basic research, genome analysis, modeling, systems biology and education.
No comments:
Post a Comment
Note: Only a member of this blog may post a comment.